Fast coalescent-based support values
Our new paper on coalescent-based branch support estimation just came out at MBE.
We introduce a fast and simple method for computing support for branches of an unrooted species tree according to the multi-species coalescent model. The main insight behind the method is simple. For any quartet of species, we can compute the frequencies of its three possible topologies in gene trees, and we can translate these frequencies to probabilities that each of the alternatives is correct. The topology with the highest frequency is the most likely species tree topology. But how much more likely is that dominant topology compared to the two alternative topologies? The answer depends on 1) the number of genes, and 2) the difference between the quartet frequency of the dominant topology and the alternatives. This relationship can be analytically derived easily by using properties of a multinomial distribution. The figure below shows an example of the relationship between a branch’s quartet support and its probability of correctness.
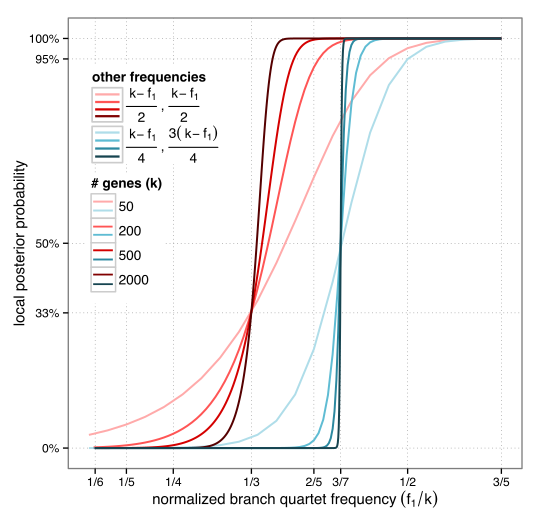
Starting with this principle, we show how all quartet frequencies can be quickly computed for each branch in a given species tree (the running time is linear in the number of species and the number of genes for each species tree branch). We further use some simplifying assumptions, which enable us to compute a branch support by summarizing the frequencies of all quartets around the branch. Perhaps our most important assumption is the “locality” assumption: for any branch, we assume all the four clusters around it are correct.
We test the new support calculation method on datasets that violate our assumptions and have high levels of gene tree estimation error. In simulation studies, we show that local posterior probabilities are remarkably accurate and give a more reliable measure of support than the standard multi-locus bootstrapping.
Our new approach fits very well within ASTRAL. ASTRAL now uses local posterior probabilities to quickly compute branch support for the species tree topology that it computes. ASTRAL can compute branch support for a dataset with 1000 genes and 1000 species in a matter of minutes on a normal laptop.
By the way, we also use computed quartet frequencies for computing branch lengths in coalescent units for internal branches of the species tree (e.g., ASTRAL’s output). This feature is also added to ASTRAL.