Visualization Techniques in Multiple Working Environments
Interactive Scientific Visualization Techniques in Multiple Working Environments
 |
 |
| CAVE |
HI-Space |
Project Overview
Visualization tools have proven to help scientists with discovery and confirmation tasks, but the tools are still inadequate for many complicated problems and are not keeping pace with advances in data generation. Furthermore, visualization tools have been optimized, in general, only for conventional desktop displays and interaction devices. This project will advance the state of the art in visualization and user interface design through research and development of novel, human-centered interactive visualization techniques designed for advanced interaction environments such as large display spaces, which allow head-tracked stereo viewing, and multimodal interaction.
Our driving application is a component of the Microbial Cell Project (MCP), a Department of Energy (DOE) program. The goal of the MCP is to understand the complex functioning of a single microbial cell, to use its capabilities for energy production and environmental cleanup. In this project, a set of prototype visualization and interaction tools will be created specifically to address some of the complicated visualization needs of MCP researchers. Our hypothesis is that human-centered environments are superior for complex data analyses, because these environments are significantly more natural and intuitive than a conventional desktop environment.
This project is a collaboration of Brown University with the Pacific Northwest National Laboratories (PNNL). Both sites are developing software for two human-centered environments, located at these institutions: the Cave at Brown University, and the Human Information Workspace (HI-Space) at PNNL. Our goal is to find out about the advantages and disadvantages of these environments when used to solve tasks from the MCP, in comparison to traditional desktop PCs or Fish Tank VR.
Results
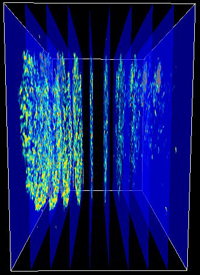 |
A cell data set consists of a stack of slices. The more slices we have, the better the 3D reconstruction. |
 |
This is what the reconstructed cell data set looks like in the Cave. It is being displayed together with another data set, which is currently in region of interest viewing mode: the system allows to work with as many data sets as the hardware can handle. Click on the image to see a little movie about how it works (MPEG4 video w/audio, 20MB, requires Quicktime). |
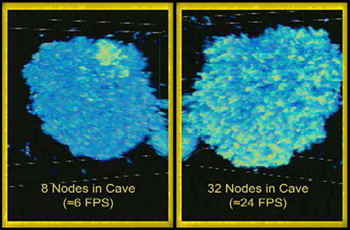 |
We ran our texture hardware based volume rendering approach in the Cave and compared the performance of different numbers of nodes. |
Click here to see what interactions the system currently supports in the Cave.
Researchers
Jürgen Schulze
Andrew Forsberg
Andries van Dam
Links
Human Centered Environments at PNNL
Human Information Workspace (HI-Space)
Microbial Cell Project (MCP)
Genomes to Life Project (GTL)
Publications
-
J.P. Schulze, A.S. Forsberg, A. Kleppe, R.C. Zeleznik, D.H. Laidlaw
"Characterizing the Effect of Level of Immersion on a 3D Marking Task"
To appear in Proceedings of HCI International, Las Vegas, July 22-27, 2005
-
R.C. Zeleznik, A.S. Forsberg, J.P. Schulze
"Look-That-There: Exploiting Gaze in Virtual Reality Interactions"
Technical Report, Brown University, Department of Computer Science, March 2005
- J.P. Schulze, A.S. Forsberg
"A Comparison of a Cave and a Fish Tank VR System for Counting Biological Features in a Volume"
Technical Report, Brown University, Department of Computer Science, March 2005
- D. Ehrens, A. Tandon, R. Balasubramanian, J.P. Schulze
"Volume Rendering with Animation of Gulf Stream Currents"
Technical Report, Brown University, Department of Computer Science, March 2005
- J.P. Schulze, A.S. Forsberg
"User Friendly Volume Data Set Exploration in the Cave"
White paper for the IEEE Visualization 2004 contest, Austin, TX, published on the conference web site, October 2004
-
J.P. Schulze, A.C. Rice
"Real-Time Volume Rendering of Four-Channel Data Sets"
In Poster Proceedings of IEEE Visualization 2004, Austin, TX
-
A.J. Cowell, R. May, N. Cramer
"The Human-Information Workspace (HI-Space): Ambient Table Top Entertainment"
In Proceedings of the 3rd International Conference on Entertainment Computing, Eindhoven, The Netherlands, September 2004, pp. 101-107
-
R. May
"Merging Ubiquitous Displays and Interactions in the Electronic and Physical Spaces"
Ubiquitous Computing 2004 workshop on Ubiquitous Display Environments, Nottingham, England, September 7, 2004
Contact
Jürgen Schulze





